In this section we will show that information on the distibution of cells ob tained at one point in the cell cycle can be generally transferred to othere places in the cell cycle.
Data from Creanor & Mitchison (1993) on protein synthesis in
fission yeast cells clearly indicate that the distribution of cell cycle times
cells is bounded on both sides and bell shaped (fig. 5), and
are best represented by the ![]() -distribution.
-distribution.
We had a problem applying our transformation methods to most of their
experiments for the number of data
points were to low in order to reliably estimate the (higher order)
derivatives with respect time necessary for our method. Only the experiments
given in figs. 9 & 10 (in Creanor & Mitchison, 1993) had
a reasonable number of data points, and we applied the methods on those
experiments only (fig. ![]() ). We observe that each method
gives rise to a considerable negative part of underlying individual output
function. The reason for this negative part is that the
distribution function for cell cycle times is least as broad as the peak in
enzyme concentration. If the width of the peak in enzyme concentration is
smaller then the width of the distribution of cell cycle times necessarily the
underlying individual output function
must have negative values. Since concentrations can not be negative
the results we obtain are flaws, and indicate that the assumption made,
that the distribution taken from events at the end of the cycle can be caried
over to the distribution of cells at the moment of the start of CDC13, is
is falsified. The distribution of cells at the moment of starting CDC13
build up has to have a smaller width to allow for positive enzyme
concentrations.
). We observe that each method
gives rise to a considerable negative part of underlying individual output
function. The reason for this negative part is that the
distribution function for cell cycle times is least as broad as the peak in
enzyme concentration. If the width of the peak in enzyme concentration is
smaller then the width of the distribution of cell cycle times necessarily the
underlying individual output function
must have negative values. Since concentrations can not be negative
the results we obtain are flaws, and indicate that the assumption made,
that the distribution taken from events at the end of the cycle can be caried
over to the distribution of cells at the moment of the start of CDC13, is
is falsified. The distribution of cells at the moment of starting CDC13
build up has to have a smaller width to allow for positive enzyme
concentrations.
An exact trace of CDC13 concentration could never have been generated from this data for we still have to decide which transformation method is the proper one. But, knowing that the information on the distribution of cell cycle times does not hold at the moment of starting CDC13 build up, we clearly cannot use any method presented here to
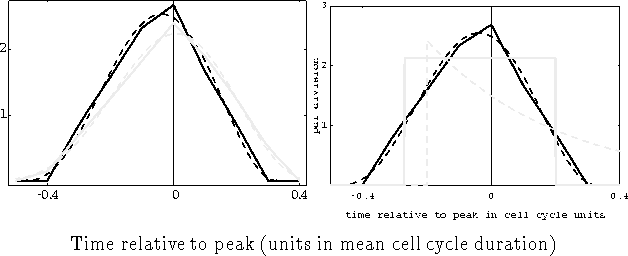
Figure 5: Distribution functions for wildtype (black line in A and B) and
cdc13 mutant (gray line in A) cells of fission yeast (From
Creanor & Mitchison (1993). A:The dashed lines are the results of fitting
the ![]() -distribution to the experimental data. B:For the wildtype
cells additional distributions were fitted: The uniform distribution (solid
gray line, and the exponenetial distribution (dashed gray line).
-distribution to the experimental data. B:For the wildtype
cells additional distributions were fitted: The uniform distribution (solid
gray line, and the exponenetial distribution (dashed gray line).